About MDAKits
What is an MDAKit?
MDAnalysis Toolkits (MDAKits) are standalone packages containing code using components from the MDAnalysis library with the aim of solving specific scientific problems or in some form enhance the functionality of the MDAnalysis core library. An MDAKit can be written by anyone and hosted anywhere.
At the discretion of their authors, MDAKits can be registered in this MDAKit registry. To do so, a MDAKit has to meet a minimum set of requirements:
The code in the package uses the MDAnalysis library
The code is open source and published under an OSI approved license
The code is versioned and provided in an accessible version-controlled repository (GitHub, GitLab, Bitbucket, etc.)
Code authors and maintainers are clearly designated
Minimal documentation is provided (what your code does, how to install it, and how to use it)
At least minimal regression tests are present; continuous integration is encouraged
The source code is installable as a standard package
It is also highly encouraged that the MDAKit also satisfies:
Information on bug reporting, user discussions, and community guidelines is made available
The code is made available on a package distribution platform (e.g. PyPi or conda-forge).
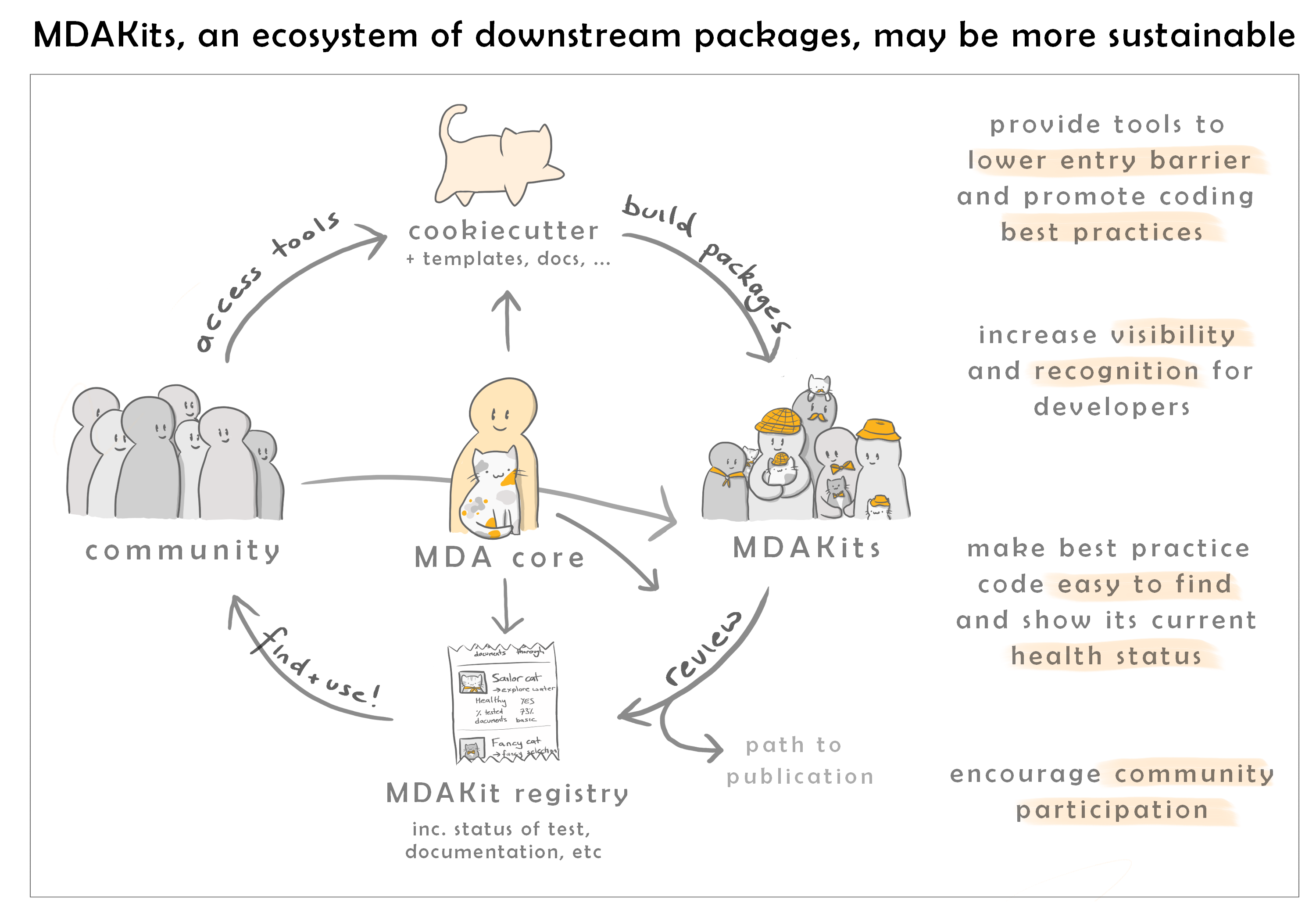
Registered MDAKits are then listed here with the intention of providing a centralised place where the community can find out more about them. Where possible, the MDAKits are also continually tested against the latest and development versions of MDAnalysis. This ensures that users and developers have an up-to-date view of the code health of an MDAKit.
The specifications are written out in detail in the MDAKits paper.
Why?
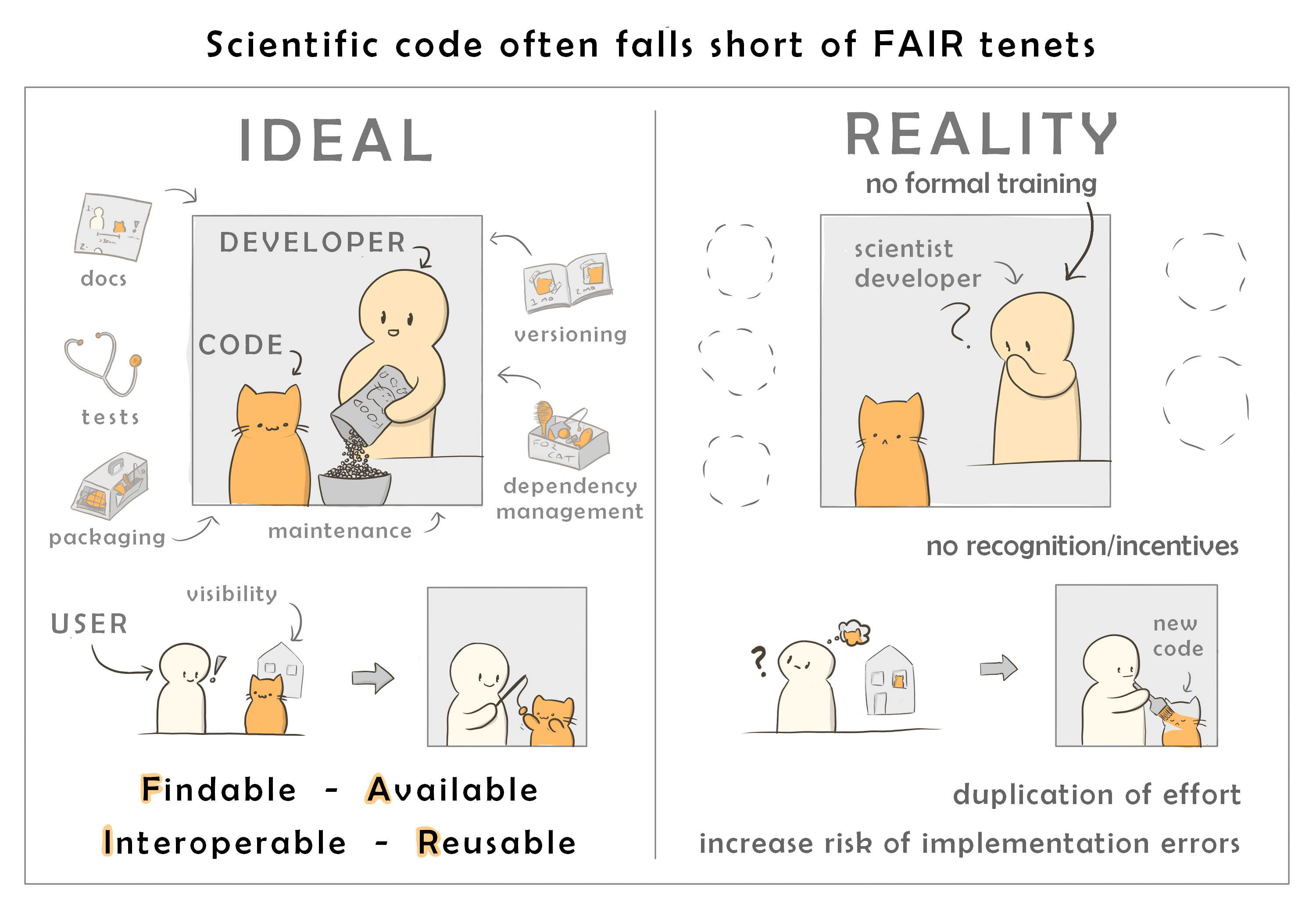
The open sharing of code that abides by the basic principles of FAIR (findability, accessibility, interoperability, and reusability) is essential to robust, reproducible, and transparent science. However, scientists typically are not supported in making the subtantial effort require to make software FAIR-compliant.
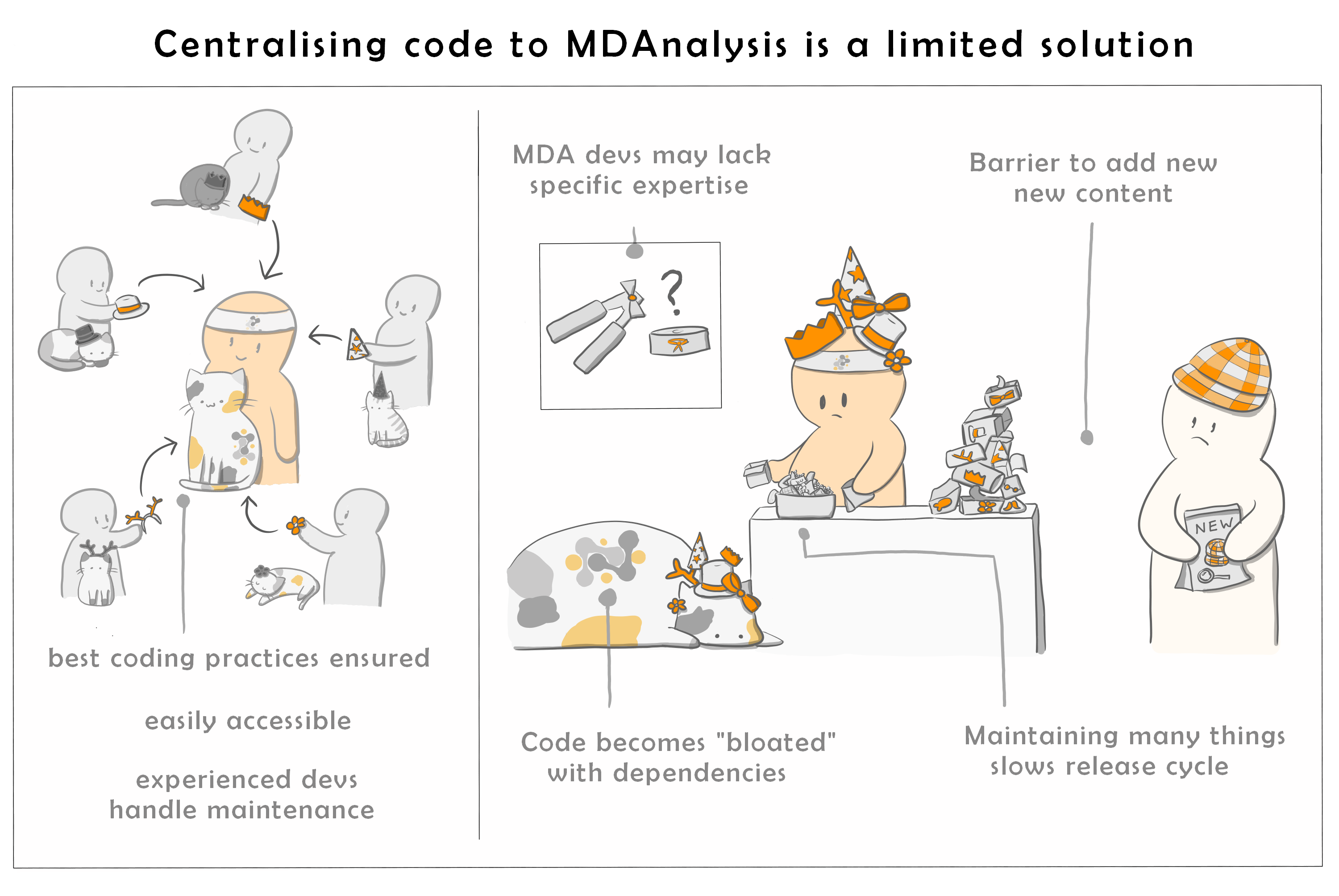
One potential option, and one that MDAnalysis has historically attempted, is to attempt to add as many methods as possible in a single centralised library. However, this is a non-ideal solution.
Our goal with MDAKits is to lower the barrier for researchers to produce FAIR software.
We wish to support developers in creating new pakages, guiding them through the process of achieving best practices and FAIR compliance. At the same time, we hope to make MDAnalysis useful to a broader community.
Read our SciPy proceedings paper!
To learn more about MDAKits and our vision for more sustainable community developed molecular simulation tools, please see our SciPy proceedings paper:
Irfan Alibay, Lily Wang, Fiona Naughton, Ian Kenney, Jonathan Barnoud, Richard J Gowers, and Oliver Beckstein. MDAKits: A framework for FAIR-compliant molecular simulation analysis . In Meghann Agarwal, Chris Calloway, and Dillon Niederhut, editors, Proceedings of the 22nd Python in Science Conference (SCIPY 2023), pages 76–84, Austin, TX, 2023. doi: 10.25080/gerudo-f2bc6f59-00a.